Team Darwin
Team Members
- Simon Goring – University of Wisconsin
- Eric Grimm – University of Minnesota
- Michael McClennen - University of Wisconsin
- Jon Nichols – Columbia University
- Mark Uhen – George Mason University
- John Wieczorek – University of California Berkeley
- Jack Williams – University of Wisconsin
Project Goals
- Crosswalk paleobiological resources (Neotoma, SESAR) onto DarwinCore standard.
- Publish Paleobiology Database and Neotoma for integration in VertNet, iDigBio, and GBIF.
Darwin Core
- Introduction - http://rs.tdwg.org/dwc/index.htm
- Quick Reference Guide (term reference) - http://rs.tdwg.org/dwc/terms/index.htm
- Biodiversity Data Standards (TDWG) - http://www.tdwg.org/
Neotoma
- Neotoma manual - http://neotoma-manual.readthedocs.io/en/latest/
- Neotoma entity relationship diagram - http://www.neotomadb.org/uploads/NeotomaDMD.pdf
Neotoma-DWC
- Initial crosswalk table - https://docs.google.com/spreadsheets/d/1mlb_VsOSps5PS4wpCw0AtmV1mV4b5pmjlBz1q_4FLUA/edit?usp=sharing
Mobilization Workflow
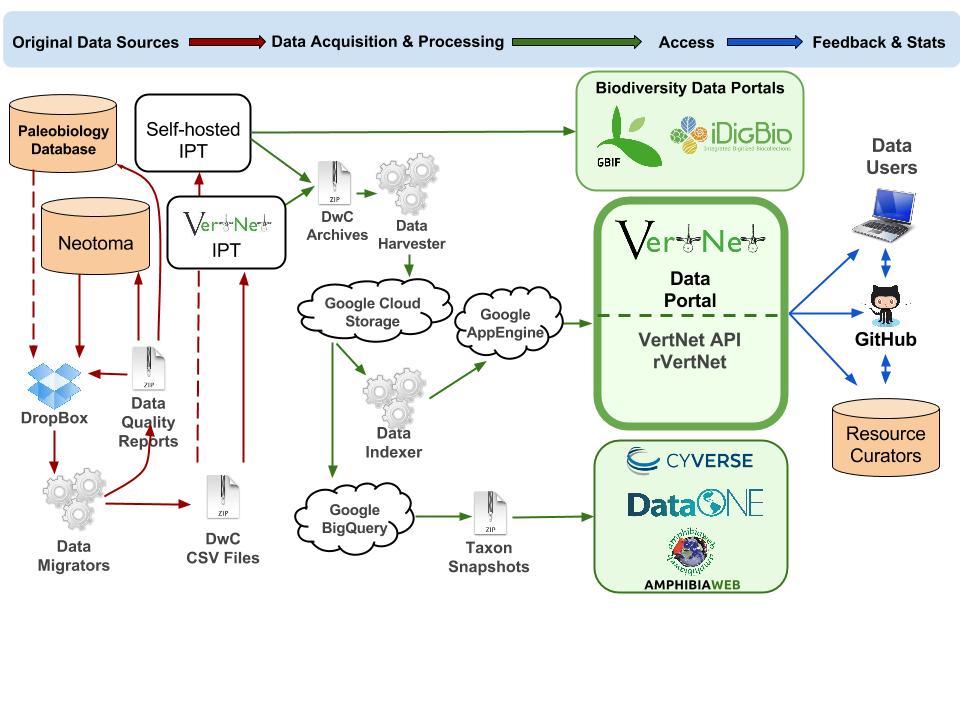
This workflow shows the steps used to make data accessible in large-scale aggregators such as VertNet, iDigBio, and GBIF using the VertNet model for data mobilization.
Workflow steps
- An original data source (e.g., Paleobiology Database, Neotoma) exports relevant data to a Dropbox folder shared with VertNet.
- VertNet adapts a custom “migrator” to the original data
- The migrator uses data cleaning and lookup routines to the most complete and correct Darwin Core output feasible from the source data.
- The migrator creates data quality and change reports as well as a Darwin Core-compliant CSV file and puts them in the shared Dropbox folder for review by the original data source curators.
- Original data source curators use reports to a) improve original data at the source or b) request improvements to the controlled vocabularies and data quality routines.
- Original data source curators share improved data via Dropbox and continue the data quality improvement cycle until there is agreement that the data set is ready for publication.
- Darwin Core CSV file is uploaded into a resource (data set and organization registered with GBIF) in an instance of the Integrated Publishing Toolkit (IPT) where it is combined with resource metadata and “published” (made openly accessible via http at the registered resource location) as a Darwin Core Archive. The IPT maybe be hosted by a third party (as in the case of Neotoma on the VertNet IPT, or self-hosted, as in the case of Paleobiology Database).
- Aggregators such as VertNet, iDigBio, and GBIF monitor IPT RSS feeds for publication events, and harvest data for inclusion or updates in their respective services.
- For VertNet, a series of steps prepare the data for inclusion in the VertNet data store as well as in periodic Taxon-based snapshots (roughly divided among Chordate classes) for archiva purposes in repositories such as CyVerse and DataONE. AmphibiaWeb harvests one of these snapshots for periodic updates to their occurrence data.
- VertNet also gathers user-provided data quality feedback and usage statistics, which it propogates as issues to anyone monitoring the data sets via their GitHub feedback repositories - one repository per IPT resource.
Workflow Diagram